MPI-VGAE: protein–metabolite enzymatic reaction link prediction
Abstract
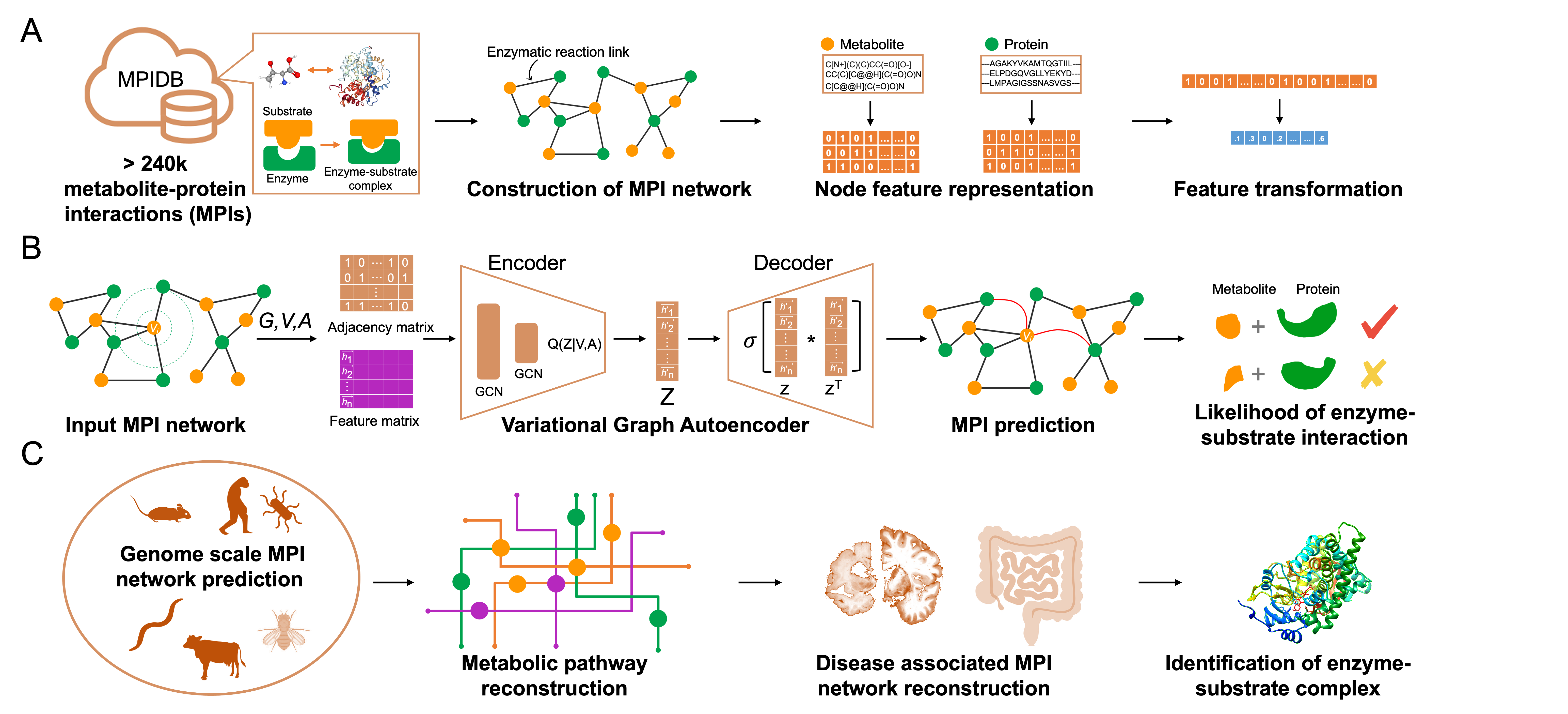
Enzymatic reactions are crucial for exploring the mechanistic function of metabolites and proteins in cellular processes and understanding the etiology of diseases. The increasing number of interconnected metabolic reactions allows the development of in silico deep learning-based methods to discover new enzymatic reaction links, expanding the landscape of the existing metabolite–protein interactome. Here, we present a Variational Graph Autoencoders (VGAE)-based framework to predict metabolite–protein interactions (MPI) across ten organisms. The MPI-VGAE predictor achieved the best predictive performance compared to other state-of-the-art methods. The MPI-VGAE framework has been applied to reconstruct MPI networks in Alzheimer’s disease and colorectal cancer and identify new enzymatic reaction links. This study highlights the potential of the MPI-VGAE framework for discovering unknown enzymatic reactions and facilitating the study of disrupted metabolisms in diseases.
